Forschung
| End of August 2016, the group of Attila Németh has stopped working at the University of Regensburg. On these sides you find a synopsis of their accomplishments as well as the new contact information of Attila Németh. The research of our group is focused on the biology of the mammalian nucleolus. On the one hand we investigate the function of the nucleolus in genome organization and establishment of nuclear architecture, on the other hand we work on the elucidation of molecular mechanisms that coordinate ribosomal gene transcription with changes in cell growth. |
| Nucleolar control of genome organization |
| Nucleolus-associated chromosomal domains (NADs) are structural and functional units of genome organization in the interphase. NADs are enriched in different gene families and certain satellite repeats, and they represent mainly a heterochromatin environment in addition to transcriptionally active ribosomal DNA. Currently we address the following questions:
|
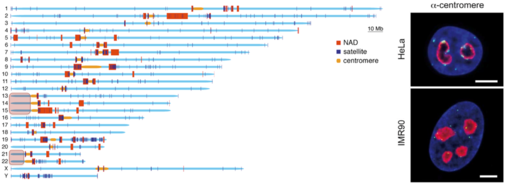 |
| Figure 1. Distribution of NADs along human chromosomes in HeLa cells. Nucleolar organizer regions are indicated with light red boxes. Confocal microscopy images on the right illustrate frequent association of centromeres with nucleoli (Németh et al., 2010). |
| Regulation of ribosomal RNA synthesis and processing |
|
We aim to answer the following questions in our ongoing projects:
|
|
|
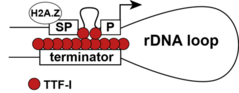
| Figure 2. Loop model of rRNA gene transcription. TTF-I binds next to the spacer promoter (SP) and the gene promoter (P) of ribosomal DNA (rDNA) and to multiple binding sites in the terminator region. Promoter-terminator interactions are mediated by TTF-I suggesting looping of active genes (Németh et al., 2008, Diermeier et al., 2014). |
Team
Attila Nemeth | curriculum vitae |
Former mentees | |
| Dillinger, Stefan | MSc/PhD student |
| Zillner, Karina | MSc/PhD student |
| Fladerer, Sebastian | MSc student |
| Porfenenko, Mikhail | MSc student |
| Filarsky, Katharina | MSc student |
| Rachow, Kathrin | MSc student |
| Fladerer, Sebastian | BSc student |
| Thies, Carina | BSc student |
| Kuttenberger, Verena | BSc student |
| Fink, Katharina | BSc student |
Attila Nemeth - Short CV
| since 2017 | Laboratory Manager - JLU Gießen (Germany), Neuropathologie |
| since 2016 | Staff Scientist - IMB Mainz (Germany), Developmental Epigenomics Group |
| 2012 - 2016 | Reader (Akademischer Oberrat, Privatdozent) - University of Regensburg (Germany), Dept. of Biochemistry III |
| 2006 – 2012 | Lecturer (Akademischer Rat) - University of Regensburg (Germany), Dept. of Biochemistry III; Habilitation in Biochemistry |
| 2007 – 2008 | Visiting Scientist - LMU Munich (Germany), Dept. of Human Genetics |
| 2002 – 2006 | Postdoc - LMU Munich (Germany), Dept. of Molecular Biology |
| 1997 – 2002 | PhD in Oncology - Semmelweis University (Budapest, Hungary) / DKFZ (Heidelberg, Germany) |
Publikationen
SCIENTIFIC ARTICLES INDEXED BY PUBMED
22) Filarsky M.*, Zillner K.*, Araya I.*, Villar-Garea A., Merkl R., Längst G.#, Németh A.# (2015) The extended AT-hook is a novel RNA binding motif. RNA Biol 12, 2015 Jul 9:0. [Epub ahead of print]
21) Zillner K., Komatsu J., Filarsky K., Kalepu R., Bensimon A., Németh A. (2015). Active human nucleolar organizer regions are interspersed with inactive rDNA repeats in normal and tumor cells. Epigenomics 7, 363-78
20) Hamdane N., Stefanovsky V., Tremblay M.G., Németh A., Paquet E., Lessard F., Sanij E., Hannan R.D. and Moss T. (2014) Conditional inactivation of Upstream Binding Factor reveals its epigenetic functions and the existence of a somatic nucleolar precursor body. PLoS Genet 10, e1004505
19) Dillinger S.*, Villar-Garea A.*, Deutzmann R., Németh A. (2014). Analysis of histone post-translational modifications from nucleolus-associated chromatin by mass spectrometry. Methods Mol Biol, 1094, 277-93
18) Németh A. (2014). Methyl-combing—single molecule analysis of DNA modifications on stretched DNA fibers. Methods Mol Biol, 1094, 233-41
17) Diermeier S., Németh A., Rehli M., Grummt I., Längst G. (2013) Chromatin-specific regulation of mammalian rDNA transcription by clustered TTF-I binding sites. PLoS Genet 9, e1003786
16) Zillner K., Filarsky M., Rachow K., Weinberger M., Längst G.#, Németh A.# (2013) Large-scale organization of ribosomal DNA chromatin is regulated by Tip5. Nucleic Acids Res 41, 5251-62.
15) Németh A.#, Perez-Fernandez J., Merkl P., Hamperl S., Gerber J., Griesenbeck J., Tschochner H.# (2013). RNA polymerase I termination: where is the end? BBA - Gene Regulatory Mechanisms 1829, 306-17. Review.
14) Hochstatter J., Hölzel M., Rohrmoser M., Schermelleh L., Leonhardt H., Keough R., Gonda T.J., Imhof A., Eick D., Längst G.#, Németh A.# (2012). Myb-binding protein 1a (Mybbp1a) regulates levels and processing of pre-ribosomal RNA. J Biol Chem 287, 24365-77.
13) Reiter A.*, Hamperl S.*, Seitz H., Merkl P., Perez-Fernandez J., Williams L., Gerber J., Németh A., Léger I., Gadal O., Milkereit P., Griesenbeck J., Tschochner H. (2012). The Reb1-homolog Ydr026c/Nsi1 is required for efficient RNA polymerase I termination in yeast. EMBO J 31, 3480-93.
12) Zillner K., Németh A. (2012). Single molecule, genome-scale analyses of DNA modifications: exposing the epigenome with next generation technologies. Epigenomics 4, 403-14. Review.
11) Felle M., Joppien S., Németh A., Diermeier S., Thalhammer V., Dobner T., Kremmer E., Kappler R., Längst G. (2011). The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res 39, 8355-65.
10) Németh A.#, Längst G.# (2011). Genome organization in and around the nucleolus. Trends Genet 27, 149-56. Review.
9) Németh A., Conesa A., Santoyo-Lopez J., Medina I., Montaner D., Péterfia B., Solovei I., Cremer T., Dopazo J., Längst G. (2010). Initial genomics of the human nucleolus. PLoS Genet 6, e1000889.
Highlighted in Nature (2010 Apr 1;464(7289):652.) and Nature Reviews Genetics (2010 May 1;11(5):314.), and selected for the „PLoS Collection: Epigenetics 2010“
8) Németh A., Längst G. (2008). Chromatin organization of active ribosomal RNA genes. Epigenetics 3, 243-5.
7) Németh A., Guibert S., Tiwari V.K., Ohlsson R., Längst G. (2008). Epigenetic regulation of TTF-I mediated promoter-terminator interactions of rRNA genes. EMBO J 27, 1255-65.
6) Espada J., Ballestar E., Santoro R., Fraga M. F., Villar-Garea A., Németh A., Lopez-Serra L., Ropero S., Aranda A., Orozco H., Moreno V., Juarranz A., Stockert J. C., Längst G., Grummt I., Bickmore W., Esteller M. (2007). Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Res 35, 2191-8.
5) Németh A., Strohner R., Grummt I., Längst G. (2004). The chromatin remodeling complex NoRC and TTF-I cooperate in the regulation of the mammalian rRNA genes in vivo. Nucleic Acids Res 32, 4091-4099.
4) Németh A., Längst G. (2004). Chromatin higher order structure: opening up chromatin for transcription. Brief Funct Genomic Proteomic 2, 334-43. Review.
3) Strohner R., Németh A., Nightingale K.P., Grummt I., Becker P.B., Längst G. (2004). Recruitment of the nucleolar remodeling complex NoRC establishes ribosomal DNA silencing in chromatin. Mol Cell Biol 24, 1791-1798.
2) Várkonyi J., Kovalszky I., Németh A., Demeter J., Raposa T. (2001). Increased risk for cancer in multiple myeloma patients and their first-degree relatives. Haematologia 31, 45-50.
1) Strohner R.*, Németh A.*, Jansa P., Hofmann-Rohrer U., Santoro R., Längst G., Grummt I. (2001). NoRC--a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J 20, 4892-4900.
* joint first authors
# joint corresponding authors
OTHER PUBLICATIONS
5) Németh A., Längst G. (2013). Chromatin organization and the mammalian nucleolus. In: Proteins of the Nucleolus (Springer), 119-48. Book Chapter.
4) Zillner K., Komatsu J., Filarsky K., Bensimon A., Németh A. (2012). Deciphering repetitive DNA methylation patterns on single DNA fibers using Epi-Combing. FEBS Journal 279, SI Supp. 1 512-3. Meeting Abstract.
3) Diermeier S., Németh A., Exler J., Längst G. (2012). Epigenomic characterization of the structure-function relation in chromatin. FEBS Journal 279, SI Supp. 1 480. Meeting Abstract.
2) Griesenbeck J., Längst G., Milkereit P., Németh A., Tschochner H. (2011). Ribosomen-Biogenese: Hierarchie oder koordiniertes Miteinander? BioSpektrum 13, 750-3.
1) Németh A., Rachow K., Hoffmeister H., Strohner R., Weinberger M., Längst G. (2009). NoRC regulates higher order rDNA structure. Cell Oncol 31, 146-7. Meeting Abstract.
Kontakt
PD Dr. Attila Németh
Laboratory Manager
Institut für Neuropathologie
Forschungslabor
Aulweg 128; 1. OG
35392 Gießen
Germany
Phone: +49 (0)641-99-30501
Fax: +49 (0) 641-99-30509
Email: Attila.Nemeth[at]patho.med.uni-giessen.de