Research
Institute for Biophysics and physical Biochemistry
Biochemistry II
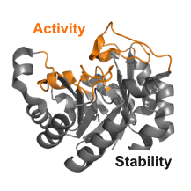
In the focus of our research are enzymes, which are elaborate proteins that catalyse cellular reactions with high specificity and efficiency.
We are using rational design and directed evolution to
- engineer the stability and catalytic activity of enzymes,
- experimentally reconstruct the natural evolution of enzymes,
- assign functions to uncharacterised enzymes,
- characterize allosteric interactions within multi-enzyme complexes
- and light-regulate enzymatic activity.
To this end, we apply a broad range of experimental and computational methods.
Our work is part of the following research clusters: