Research
Biochemistry II
Dr. Andrea Kneuttinger
Photoxenoprotein Engineering
The integration of a sensitive photoswitch allows to precisely control where and when the enzyme is active. This spatio-temporal control of enzyme activity is highly desirable, for example:
- to study functional properties of enzymes either in vitro or in the cell,
- to fine-tune enzyme cascades in industrial biocatalysis, which facilitates sustainable and greener production of various compounds,
- and to reduce side-effects of biotherapeutic enzymes, such as in chemotherapy, among other things.
For this, we specialize on the method of photoxenoprotein engineering [16], in which either photocaged or photoswitchable unnatural amino acids (UAAs) are incorporated into an enzyme through genetic code expansion. This technique achieves either irreversible activation or reversible switching between “off” and “on” states (Figure 1).
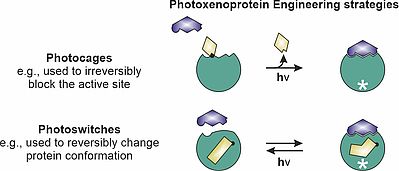
Figure 1: Irreversible activation using photocaged UAAs and reversible switching between "off" and "on" states using photoswitchable UAAs. * = enzyme activity "on".
Our Goals
While photoxenoprotein engineering is highly versatile it is still very "young" , which opens up plenty of possibilities to apply, investigate and further develop this photocontrol method. Hence, our goals focus on:
|
Our Toolbox
Enzymes
Our previous success of photocontrolling enzyme activity with unnatural amino acids correlated with the disruption of allostery and essential motions in the enzyme of interest [10, 12, 13]. Therefore, two of our projects focus on the two model enzyme complexes imidazole-glycerol phosphate synthase and tryptophan synthase. However, in recent years we have extended our scope of research to non-allosteric enzymes that are of interest for cell studies, industrial biocatalysis and biotherapy.
Genetic code expansion
For the efficient incorporation of our light-sensitive unnatural amino acids we employ the amber stop codon suppression technology, which is outlined in Figure 2. An orthogonal aminoacyl-tRNA synthetase (O-aaRS) is redesigned for binding the unnatural amino acid (UAA) and coupling it to an orthogonal tRNA (O-tRNA). At the ribosome, the anticodon of the O-tRNA binds to a reprogrammed stop codon allowing the nascent protein chain to react with the UAA. The result of this reprogrammed protein synthesis is a protein bearing an UAA at a specified position.
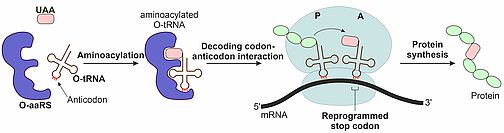
Figure 2: Incorporation of an unnatural amino acid into a protein.
Protein Engineering
Every marginal function of an enzyme can be improved by protein engineering. We use a rational design approach with low-throughput screening assays to establish initial photocontrol in our enzymes of interest. In order to improve the efficiency and power of photocontrol in these initial systems, we moreover apply various directed evolution strategies with medium- to high-throughput activity screenings.
Photoxenoprotein characterization
Extended enzymological, biophysical and biochemical studies constitute the core of our research. This primarily includes the analysis of enzyme activity. For this we establish continuous activity measurements using coupled enzyme assays, and measure steady-state kinetics of pre- and post-irradiation states as well as the return to the steady-state after irradiation of the assay (dubbed “direct photocontrol”)[17]. Furthermore, we characterize our photoxenoproteins regarding, e.g., ligand binding, protein complex formation and structural changes, for which we benefit from a large methodological framework including absorbance and fluorescence spectroscopy, analytical size exclusion chromatography, circular dichroism spectroscopy, isothermal titration calorimetry, surface plasmon resonance, stopped-flow spectrometry and protein crystallization.
Engineering of Photocontrol in Enzymes
Group of
Dr. Andrea Kneuttinger
Universitätsstrasse 31
93053 Regensburg
Email
Phone: +49-941-943 7439
Room: E3_1.315