Topic
Summary
The eukaryotic genome is accommodated by the structure of chromatin. Chromatin is defined as the entity composed of the DNA – encoding the genetic information – and associated proteins. Chromatin controls the access of protein factors to the DNA and is thus an important determinant for many essential processes, such as transcription, replication, and DNA-repair. Chromatin is highly dynamic changing in structure and composition according to the functional state of genomic loci. To understand nuclear processes it is necessary to gain insight in the molecular nature of chromatin and the mechanisms involved in its establishment, maintenance and alteration.
We are working on the analysis of chromatin structure and function with a special focus on chromatin alterations associated with the process of transcription. We chose the yeast model system, because it allows easy genetic manipulation and straight-forward biochemistry. Furthermore, we concentrate our efforts on two genomic loci for which a robust chromatin transition had been observed when they switch their transcriptional state. The choice of the RNA Pol II dependent PHO5 gene and the ribosomal DNA locus, containing genes transcribed by RNA Pol I and III enables comparative analysis of chromatin transcribed by the three different eukaryotic RNA polymerases. The final goal of our studies is to derive a complete molecular description of chromatin in different transcriptional states.
Below you find more details about our work on rDNA chromatin, PHO5 chromatin and the isolation of native chromosomal domains.
| rDNA Chromatin |
Actively transcribed ribosomal RNA genes are virtually free of histone molecules and bound by the HMG box protein Hmo1
In all eukaryotes the structural RNAs forming the ribosome (rRNAs) are transcribed from multicopy genes, clustered or spread over the entire genome. In the yeast S. cerevisiae, the rRNA genes are present at a single genomic location on the right arm of chromosome XII (Figure 1). The rDNA locus consists of 150 to 200 tandemly repeated transcription units. Each of the transcription units contains the RNA Polymerase III transcribed 5S rRNA gene and the RNA Polymerase I transcribed 35S rRNA gene. In actively dividing yeast cells rRNA synthesis by RNA Polymerase I is responsible for almost 70% of the total transcription in the yeast cell.
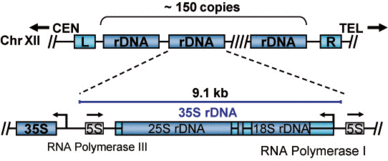
Figure 1: Schematic representation of the yeast rDNA locus. The position of the rDNA repeat cluster on chromosome XII including the left (L) and the right (R) flanking regions with respect to centromere (CEN) and telomere (TEL) is shown. Each rDNA repeat consists of the Pol I transcribed 35S rRNA gene (precursor for the 18S, 5.8S, and 25S rRNAs) and the RNA Polymerase III transcribed 5S rRNA gene. Arrows mark the transcription start sites and direction.
Apart from its dominant role in cellular transcription the rDNA locus is particular with regard to chromatin structure: Two populations of actively transcribed and transcriptional inactive 35S rRNA genes co-exist in all eukaryots investigated so far (reviewed in Hamperl et al., 2013). These two transcriptional states reside in distinct chromatin structures (Figure 2). It has been a long-lasting question why cells keep a fraction of the 35S rRNA genes in a transcriptionally inactive state. Work from Kobayashi and co-workers suggested that maintenance of this fraction is important for genome stability under certain conditions (Ide et al., 2010). Nevertheless, the molecular nature of the different chromatin states remains still largely unknown. Whereas evidence had been provided that actively transcribed rRNA genes are virtually nucleosome free, more recent data suggested that RNA Polymerase I transcribes a nucleosomal template in vivo (Jones et al., 2007).
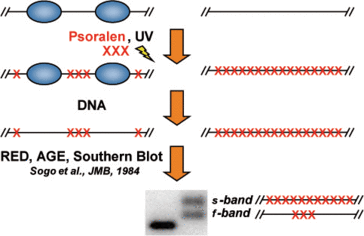
Figure 2: Two chromatin states exist within the coding region of ribosomal rRNA genes. Yeast cells are incubated in the presence of a DNA intercalating agent, Psoralen. Upon UV irradiation DNA-bound Psoralen forms a covalent bond between the two single strands linking them closely together. DNA is isolated, subjected to restriction enzyme digestion (RED), separated by agarose gel electrophoresis (AGE) and analyzed in a Southern Blot. If a probe specific for the coding regions of rRNA genes is used two distinct fragments with different migrating with mobility in the agarose gel are observed in DNA samples derived from Psoralen treated cells (+), but not in non-treated controls (-). According to their mobility they are termed slow (s-) and fast (f-) migrating band. These DNA fragments are identical in their sequence, but differ largely in their degree of Psoralen incorporation.
To investigate rDNA chromatin we have applied a technique called Chromatin Endogenous Cleavage (ChEC) (Schmid et al., 2004). In such experiments the genomic locus of a factor of interest is modified in a way that it is expressed as a fusion protein carrying a C-terminal Micrococcal nuclease (MNase). MNase is a Calcium-dependent endonuclease with only little sequence specificity. The fusion proteins are inactive in the living cell, because a very low Calcium concentration is maintained in cytoplasm and nucleoplasm. At a desired time point factors bound to their respective genomic interaction sites are covalently linked to the DNA by the addition of Formaldehyde. After isolation of nuclei Calcium is added and the MNase fusion proteins cleave the DNA in the proximity of their interaction site. The cleavage events can then be monitored and mapped with high precision to different genomic locations using indirect endlabeling Southern Blot analysis (Figure 3).
Using the ChEC technique in combination with Psoralen crosslinking analysis we could demonstrate, that actively transcribed rRNA genes are largely devoid of histone molecules but are bound by the HMG box protein Hmo1 (Figure 4) (Merz et al., 2008). This finding was consistent with the interpretation of early psoralen crosslinking analyses (Conconi et al., 1989; Dammann et al., 1993), but was not compatible with the idea that RNA Polymerase I transcribes a nucleosomal template (Jones et al., 2007).
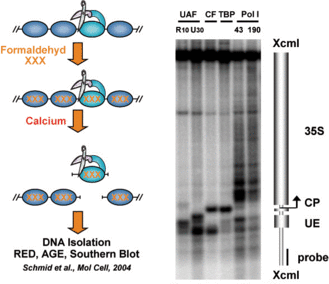
Figure 3: ChEC analysis of RNA Polymerase I and associated transcription factors at the yeast 35S rDNA sequence. ChEC was performed with nuclei isolated from yeast strains expressing the MNase fusion protein indicated on top of each panel. Isolated nuclei were incubated in calcium-containing buffer at 30°C for 30 minutes. DNA was isolated, digested with XcmI, separated in a 1% agarose gel and analyzed in a Southern Blot by indirect end-labeling with a probes visualizing a 4.9kb fragment of promoter and coding region of the 35S rRNA gene. A cartoon of the 35S rRNA gene with the corresponding transcription start sites, and the location of the probe used for indirect end-labeling are depicted on the right. CP and UE indicate the positions of core promoter and upstream element, of the 35S rDNA. UAF, upstream activating factor (including R10, Rrn10, and U30, Uaf30 subunits); CF, core factor; PolI, RNA polymerase I (including 43, Rpa43, and 190, Rpa190 subunits).
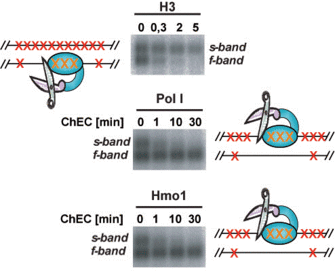
Figure 4: Hmo1 and Histone H3 associate with different states of yeast rDNA chromatin. ChEC was performed with nuclei isolated from yeast strains expressing the MNase fusion protein indicated on top of each panel. Isolated nuclei were incubated in calcium-containing buffer at 30°C for the minutes depicted above each lane. After ChEC nuclei were treated with psoralen, and DNA was isolated. The genomic DNA was digested with EcoRI, separated in a 0.8% agarose gel and analyzed in a Southern Blot with a probe detecting an EcoRI fragment within the coding region of the 35S rRNA gene.
Establishment and maintenance of 35S rRNA gene chromatin states
To investigate the cellular mechanisms being important for establishment and maintenance of 35S rRNA gene chromatin states we have studied the chromatin dynamics in course of the cell cycle. We found that the fraction of psoralen accessible (open) rRNA genes varied with the cell cycle. Replication was required to convert rRNA genes into the psoralen inaccessible (closed) chromatin state, whereas RNA Pol I transcription was needed to establish the open chromatin structure. Thus, arrest at various cell cycle stages led to opening of rRNA genes, while shut-down of RNA Pol I transcription interferred with this transition. This suggests a model in which the opposed action of replication and transcription leads to de novo establishment of chromatin states in each cell cycle (Figure 5, (Wittner et al., 2011)).
Interestingly, RNA Pol I transcription was not needed to maintain the open chromatin structure in the absence of replication. Instead, we observed conversion of rRNA genes to the closed chromatin state in the absence of the HMG box protein Hmo1 under these conditions. These findings indicate that Hmo1 stabilizes the open rRNA gene chromatin state, presumably by interferring with replication independent nucleosome assembly (Figure 5, (Wittner et al., 2011)).
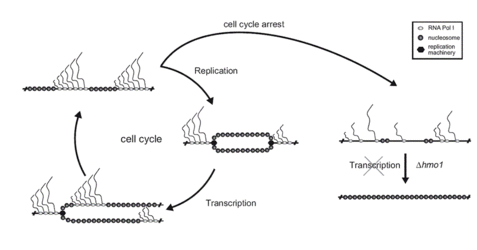
Figure 5: Establishment and maintenance of 35S rRNA chromatin states. In dividing yeast cells, the ratio of open and closed rRNA gene chromatin states is the result of a dynamic equilibrium of replication-dependent nucleosome deposition and Pol I transcription-dependent nucleosome removal. Cell-cycle arrest and thus inhibition of replication lead to complete opening of rRNA genes, provided that Pol I transcription is ongoing. In the absence of Pol I transcription, the HMG box protein Hmo1 is required for the maintenance of open rRNA gene chromatin, presumably interfering with replication-independent nucleosome assembly.
PHO5 chromatin |
Nucleosome removal from a transcriptional activated promoter in vivo
When yeast cells are starved by phosphate a signal transduction pathway is activated resulting in the expression of a subset of PHO genes involved in phosphate metabolism (Oshima, 1997). PHO5 encodes for a secreted acidic phosphatase and its promoter has served a paradigm for a defined chromatin transition upon transcriptional activation (Svaren and Horz, 1997). In the course of induction the promoter region, occupied by regularly spaced nucleosomes in repressing conditions, becomes highly accessible (Figure 6). The nature of this chromatin transition has first been interpreted as the removal of nucleosomes (Almer et al., 1986), but had become uncertain after the discovery of enzymatic activities acting on chromatin and producing altered (accessible) states of nucleosomes in vitro.

Figure 6: Structural changes in PHO5 promoter chromatin upon transcriptional activation and structural integrity of PHO5 chromatin upon purification. Restriction endonuclease accessibility of PHO5 promoter DNA in isolated nuclei and in purified chromatin circles. Nuclei isolated from yeast strains repressed (R) and activated (A) in PHO5 expression, were digested with 50 or 200 U of the indicated restriction endonuclease (Chromosome). Affinity purified PHO5 chromatin circles were digested with 5 or 20 U of the indicated restriction endonuclease (Gene circle, Promoter circle). After extraction DNA was linearized (positions marked by asterisks) and analyzed in a Southern blot with the probe indicated. An arrowhead on the left points to the position of uncut DNA. A schematic representation of the PHO5 promoter is given on the top. Ovals represent positioned nucleosomes (-1 to -4) on the promoter under repressing conditions. Black dots represent transcription factor binding sites UASp1 (U1), and UASp2 (U2). A square represents the TATA box (T).
A combination of in vivo observations and in vitro experiments with purified chromatin circles obtained by the technique described below enabled us to determine the fate of nucleosomes upon transcriptional activation: Using four different approaches employing topology, sedimentation, limit nuclease and formaldehyde cross-linking analyses we could unambiguously demonstrate that nucleosomes are lost from the activated PHO5 promoter in vivo (Boeger et al., 2003). Additionally, we analyzed transcription factor binding to the PHO5 promoter in vivo, using ChEC. The results demonstrated that nucleosomes effectively interfere with the binding of Pho4 and other critical transcription factors to regulatory sequences of the PHO5 promoter (Mao et al., 2011). Furthermore we could uncover the mechanism by which nucleosomes are removed from the PHO5 promoter. Our results rule out a sliding mechanism and argue for nucleosome disassembly upon activation of PHO5 in vivo (Boeger et al., 2004).
In continuation of the above studies, we combined biochemical analyses of the chromatin transition at the transcriptionally induced PHO5 promoter with mathematical modeling based on a small number of simple assumptions (Boeger et al., 2008). We showed that random removal and reformation of promoter nucleosomes can account for stochastic and kinetic properties of PHO5 expression. Our analysis suggests that the disassembly of promoter nucleosomes is rate limiting for PHO5 expression.
Purification and analysis of native chromatin domains |
Affinity purification of specific chromatin segments from chromosomal loci in the yeast Saccharomyces cerevisiae
To isolate natural chromatin we have employed an approach developed by Gartenberg and co-workers, using site-specific recombination and differential centrifugation (Ansari et al., 1999): Yeast chromosomes are modified by flanking an endogenous locus with recognition sites for R recombinase from Zygosaccharomyces rouxii. The modified yeast cells are then transformed with a plasmid containing the R coding sequence under the control of an galactose inducible promoter. In the presence of galactose, R is expressed and excises the flanked locus as a circle. A simple differential centrifugation protocol allows separating the chromatin circles from the bulk of genomic DNA and 90% of the total cellular protein. At this point the preparation of chromatin circles is still crude and not a suitable substrate for biochemical experiments. We have extended the method by further affinity purification (Griesenbeck et al., 2003; Griesenbeck et al., 2004). For this purpose we included a LEX A-binding cluster juxtaposed to the recombinase recognition sites in an orientation that makes it part of the excised circle (Figure 7). We also developed a recombinant adaptor fusion protein that brings together the bacterial LexA protein with a tandem affinity purification (TAP) tag (Rigaut et al., 1999), binding as well to the LEX A cluster incorporated in the chromatin circles as to two different affinity supports (Figure 7).
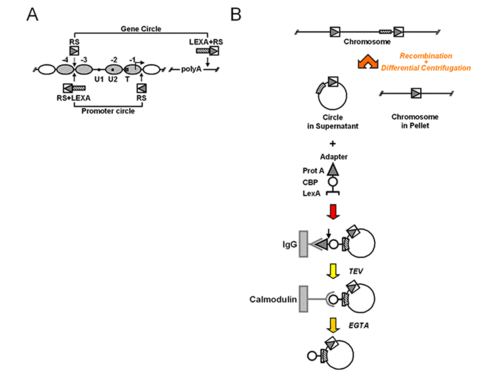
Figure 7: Targeting and isolation of a chromosomal domain. A) Genetic manipulations at the chromosomal PHO5 locus. Arrowheads point to the sites where recognition elements for R recombinase from Zygosaccharomyces rouxii (RS) and LexA operator cluster (LEXA) were inserted into the genomic locus. PHO5 domains released by homologous recombination are indicated (Gene circle, Promoter circle). B) Flow chart for recombination, differential centrifugation and affinity purification. A genetically modified chromosomal locus is excised in circular form in vivo upon induction of a site-specific recombinase. The chromatin circle carrying the tagged chromosomal domain is separated from the bulk chromatin by differential centrifugation. A recombinant adaptor molecule co-expressed in the yeast cells binds via its N-terminal LexA part (bracket) to the LexA operator cluster within the circle sequence. The adaptor molecule then bridges between the chromatin circles and two different affinity supports (IgG-agarose, Calmodulin-agarose) via a C-terminal Protein A epitope (triangle) and Calmodulin binding peptide (CBP, circle). The adaptor molecule chromatin circle complex can be released from the solid supports by proteolytic cleavage with recombinant TEV protease, and by addition of EGTA, respectively.
Using this technique we can enrich a single copy gene 180,000-fold over every other genomic locus. After the TAP procedure the chromatin circles are free from nucleases, proteases and chromatin remodeling activities. Most importantly the chromatin structure of the isolated material conserves important features of the chromatin in vivo (Figure 6). Thus, the yeast PHO5 gene could be isolated from cells grown in activating and repressing conditions preserving the chromatin structure characteristic for the respective transcriptional state (Boeger et al., 2003; Griesenbeck et al., 2003). More recently, we were able to determine the protein composition and posttranslational modification state of isolated rDNA and PHO5 chromatin domains by MS analysis and to obtain structural information by EM analyses (Hamperl et al., 2014a, Hamperl et al., 2014b). This may open the way to characterize chromatin at every chromosomal locus in molecular detail at least in yeast.
References
Almer, A., Rudolph, H., Hinnen, A., and Horz, W. (1986). Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. Embo J 5, 2689-2696.
Ansari, A., Cheng, T. H., and Gartenberg, M. R. (1999). Isolation of selected chromatin fragments from yeast by site-specific recombination in vivo. Methods 17, 104-111.
Boeger, H., Griesenbeck, J., and Kornberg, R. D. (2008). Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell 133, 716-726.
Boeger, H., Griesenbeck, J., Strattan, J. S., and Kornberg, R. D. (2003). Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell 11, 1587-1598.
Boeger, H., Griesenbeck, J., Strattan, J. S., and Kornberg, R. D. (2004). Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol Cell 14, 667-673.
Conconi, A., Widmer, R. M., Koller, T., and Sogo, J. M. (1989). Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57, 753-761.
Dammann, R., Lucchini, R., Koller, T., and Sogo, J. M. (1993). Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res 21, 2331-2338.
Griesenbeck, J., Boeger, H., Strattan, J. S., and Kornberg, R. D. (2003). Affinity purification of specific chromatin segments from chromosomal loci in yeast. Mol Cell Biol 23, 9275-9282.
Griesenbeck, J., Boeger, H., Strattan, J. S., and Kornberg, R. D. (2004). Purification of defined chromosomal domains. Methods Enzymol 375, 170-178.
Hamperl, S., Brown, C.R., Garea, A.V., Perez-Fernandez, J., Bruckmann, A., Huber, K., Wittner, M., Babl, V., Stoeckl, U., Deutzmann, R., Boeger, H., Tschochner, H., Milkereit, P., and Griesenbeck, J. (2014a). Compositional and structural analysis of selected chromosomal domains from Saccharomyces cerevisiae. Nucleic Acids Res.
Hamperl, S., Brown, C.R., Perez-Fernandez, J., Huber, K., Wittner, M., Babl, V., Stöckl, U., Boeger, H., Tschochner, H., Milkereit, P., and Griesenbeck, J. (2014b). Purification of specific chromatin domains from single-copy gene loci in Saccharomyces cerevisiae. Methods Mol. Biol. Clifton NJ 1094, 329–341.
Hamperl, S., Wittner, M., Babl, V., Perez-Fernandez, J., Tschochner, H., and Griesenbeck, J. (2013). Chromatin states at ribosomal DNA loci. Biochim. Biophys. Acta 1829, 405–417.
Ide, S., Miyazaki, T., Maki, H., and Kobayashi, T. (2010). Abundance of ribosomal RNA gene copies maintains genome integrity. Science 327, 693-696. Jones, H. S., Kawauchi, J., Braglia, P., Alen, C. M., Kent, N. A., and Proudfoot, N. J. (2007). RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat Struct Mol Biol 14, 123-130.Mao, C., Brown, C. R., Griesenbeck, J., and Boeger, H. (2011). Occlusion of regulatory sequences by promoter nucleosomes in vivo. PLoS ONE 6, e17521.
Merz, K., Hondele, M., Goetze, H., Gmelch, K., Stoeckl, U., and Griesenbeck, J. (2008). Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev 22, 1190-1204.
Oshima, Y. (1997). The phosphatase system in Saccharomyces cerevisiae. Genes Genet Syst 72, 323-334.
Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17, 1030-1032. Schmid, M., Durussel, T., and Laemmli, U. K. (2004). ChIC and ChEC; genomic mapping of chromatin proteins. Mol Cell 16, 147-157. Svaren, J., and Horz, W. (1997). Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem Sci 22, 93-97.Wittner, M., Hamperl, S., Stöckl, U., Seufert, W., Tschochner, H., Milkereit, P., and Griesenbeck, J. (2011). Establishment and maintenance of alternative chromatin States at a multicopy gene locus. Cell 145, 543-554.
team
Current members
 | Joachim Griesenbeck, Principal investigatoremail: joachim.griesenbeck (at) ur.de |
 | Kristin Hergert, Technical assistantemail: kristin.hergert (at) ur.de |
 | Philipp Milkereit, Principal investigatoremail: philipp.milkereit (at) ur.de |
 | Sebastian Kruse, PhD studentemail: sebastian.kruse (at) ur.de |
 | Herbert Tschochner, Principal investigatoremail: herbert.tschochner (at) ur.de |
Alumni
| Name | Letzte/Gegenwärtige Adresse |
| Dr. Jochen Abel | Bristol-Myers Squibb GmbH & Co, Munich, Germany |
| Dr. Virginia Babl | Novartis, Herzogenaurach, Germany |
| Dr. Christopher Dächert | Max von Pettnkofer-Institut, Munich, Germany |
| Dr. Max Felle | BASF SE, Ludwigshafen, Germany |
| Dr. Annika Frauenstein | Max-Planck-Institut für Biochemie, Munich, Germany |
| Katharina Gmelch | Maristen-Gymnasium Furth, Germany |
| Dr. Hannah Seitz (geb. Götze) | Evotech, Hamburg, Germany |
| Dr. Stephan Hamperl | Helmholtz Zentrum, Munich, Germany |
| Prof. Maria Hondele | Biozentrum, University of Basel, Switzerland |
| Dr. Julia Mändl | Last address: DKFZ, Heidelberg, Germany |
| Andreas Maier | Last address:Technische Universität München, Garching, Germany |
| Dr. Katharina Merz | STRABAG, Spital/Drau, Austria |
| Christopher Schächner | Rentschler Biopharma SE, Laupheim, Germany |
| Dr. Thomas Wild | Last adress: University of Kopenhagen, Denmark |
| Dr. Manuel Wittner | Boehringer Ingelheim Pharma GmbH, Biberach/ Riß |
Publications
Selected publications
Daiß, J.L., Griesenbeck, J., Tschochner, H., and Engel, C. (2023). Synthesis of the ribosomal RNA precursor in human cells: mechanisms, factors and regulation. Biological chemistry. https://doi.org/10.1515/hsz-2023-0214.
Merkl, P.E., Schächner, C., Pilsl, M., Schwank, K., Hergert, K., Längst, G., Milkereit, P., Griesenbeck, J., and Tschochner, H. (2022). Analysis of Yeast RNAP I Transcription of Nucleosomal Templates In Vitro. Methods in molecular biology (Clifton, N.J.) 2533, 39-59. https://doi.org/10.1007/978-1-0716-2501-9_3.
Schächner, C., Merkl, P.E., Pilsl, M., Schwank, K., Hergert, K., Kruse, S., Milkereit, P., Tschochner, H., and Griesenbeck, J. (2022). Establishment and Maintenance of Open Ribosomal RNA Gene Chromatin States in Eukaryotes. Methods in molecular biology (Clifton, N.J.) 2533, 25-38. https://doi.org/10.1007/978-1-0716-2501-9_2.
Merkl, P.E., Pilsl, M., Fremter, T., Schwank, K., Engel, C., Längst, G., Milkereit, P., Griesenbeck, J., and Tschochner, H. (2020). RNA polymerase I (Pol I) passage through nucleosomes depends on Pol I subunits binding its lobe structure. J. Biol. Chem. 295, 4782-4795. https://doi.org/10.1074/jbc.RA119.011827.
Hannig, K., Babl, V., Hergert, K., Maier, A., Pilsl, M., Schächner, C., Stöckl, U., Milkereit, P., Tschochner, H., Seufert, W., Griesenbeck, J., (2019). The C-terminal region of Net1 is an activator of RNA polymerase I transcription with conserved features from yeast to human. PLoS Genet. 15, e1008006. ![]()
Hermans, N., Huisman, J.J., Brouwer, T.B., Schächner, C., van Heusden, G.P.H., Griesenbeck, J., van Noort, J., (2017). Toehold-enhanced LNA probes for selective pull down and single-molecule analysis of native chromatin. Sci Rep 7, 16721.![]()
Babl, V., Stöckl, U., Tschochner, H., Milkereit, P., Griesenbeck, J., (2015). Chromatin Endogenous Cleavage (ChEC) as a Method to Quantify Protein Interaction with Genomic DNA in Saccharomyces cerevisiae. Methods Mol. Biol. 1334, 219–232.
Brown, C.R., Eskin, J.A., Hamperl, S., Griesenbeck, J., Jurica, M.S., Boeger, H., (2015). Chromatin structure analysis of single gene molecules by psoralen cross-linking and electron microscopy. Methods Mol. Biol. 1228, 93–121.
Hamperl, S., Brown, C.R., Perez-Fernandez, J., Huber, K., Wittner, M., Babl, V., Stöckl, U., Boeger, H., Tschochner, H., Milkereit, P., and Griesenbeck, J. (2014b). Purification of specific chromatin domains from single-copy gene loci in Saccharomyces cerevisiae. Methods Mol. Biol. Clifton NJ 1094, 329–341.
Hamperl, S., Brown, C.R., Garea, A.V., Perez-Fernandez, J., Bruckmann, A., Huber, K., Wittner, M., Babl, V., Stoeckl, U., Deutzmann, R., Boeger, H., Tschochner, H., Milkereit, P., and Griesenbeck, J. (2014a). Compositional and structural analysis of selected chromosomal domains from Saccharomyces cerevisiae. Nucleic Acids Res. ![]()
Tremblay, M., Charton, R., Wittner, M., Levasseur, G., Griesenbeck, J., and Conconi, A. (2013). UV light-induced DNA lesions cause dissociation of yeast RNA polymerases-I and establishment of a specialized chromatin structure at rRNA genes. Nucleic Acids Res. ![]()
Hamperl, S., Wittner, M., Babl, V., Perez-Fernandez, J., Tschochner, H., and Griesenbeck, J. (2013). Chromatin states at ribosomal DNA loci. Biochim. Biophys. Acta 1829, 405–417.
Griesenbeck, J., Wittner, M., Charton, R., and Conconi, A. (2012). Chromatin endogenous cleavage and psoralen crosslinking assays to analyze rRNA gene chromatin in vivo. Methods Mol. Biol. 809, 291–301.
Lorch, Y., Griesenbeck, J., Boeger, H., Maier-Davis, B., and Kornberg, R. D. (2011). Selective removal of promoter nucleosomes by the RSC chromatin-remodeling complex. Nat. Struct. Mol. Biol 18, 881-885. ![]()
Wittner, M., Hamperl, S., Stöckl, U., Seufert, W., Tschochner, H., Milkereit, P., and Griesenbeck, J. (2011). Establishment and maintenance of alternative chromatin States at a multicopy gene locus. Cell 145, 543-554. ![]()
Mao, C., Brown, C. R., Griesenbeck, J., and Boeger, H. (2011). Occlusion of regulatory sequences by promoter nucleosomes in vivo. PLoS ONE 6, e17521. ![]()
Goetze, H., Wittner, M., Hamperl, S., Hondele, M., Merz, K., Stoeckl, U., and Griesenbeck, J. (2010). Alternative chromatin structures of the 35S rRNA genes in Saccharomyces cerevisiae provide a molecular basis for the selective recruitment of RNA polymerases I and II. Mol. Cell. Biol 30, 2028-2045. ![]()
Merz, K., Hondele, M., Goetze, H., Gmelch, K., Stoeckl, U., and Griesenbeck, J. (2008). Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev 22, 1190-1204. ![]()
Boeger, H., Griesenbeck, J., and Kornberg, R. D. (2008). Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell 133, 716-726. ![]()
Griesenbeck, J., Boeger, H., Strattan, J. S., and Kornberg, R. D. (2004). Purification of defined chromosomal domains. Methods Enzymol 375, 170-178.
Boeger, H., Griesenbeck, J., Strattan, J. S., and Kornberg, R. D. (2004). Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol Cell 14, 667-673.
Griesenbeck, J., Boeger, H., Strattan, J. S., and Kornberg, R. D. (2003). Affinity purification of specific chromatin segments from chromosomal loci in yeast. Mol Cell Biol 23, 9275-9282. ![]()
Boeger, H., Griesenbeck, J., Strattan, J. S., and Kornberg, R. D. (2003). Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell 11, 1587-1598.
All relevant publications in PubMed.
Funding
-
since 2024
DFG Individual Grant GR 1706/7-1 "The role of RNA polymerase I lobe-binding subunits in regulation of transcription, rRNA proofreading and quality control of ribosome biosynthesis"
-
2011 - 2023
DFG collaborative research Center SFB960 at the University of Regensburg: Ribosome formation: principles of RNP biogenesis and control of their function
Project A5: Compositional analysis of ribosomal RNA gene chromatin in disinct transcriptional states
-
2009 - 2018
Mobility programme accompanied by the Bavarian Research Alliance GmbH as an assignment from the Bavarian State Chancellery for initial networking with the group of Antonio Conconi, Université de Sherbrooke, Canada
![]()
-
2007 - 2011
DFG research unit FOR1068 at the University of Regensburg: From the chromatin to the ribosome - Regulation and mechanisms of ribosome biogenesis.
Project A5: Compositional and posttranslational covalent modification of chromatin in distinct transcriptional states of the ribosomal DNA

-
2006 - 2016
Project accompanied by the Bavaria California Technology Center together with the group of Hinrich Boeger, UCSC, USA
![]()
-
2006 - 2008
CNRS - PROJET INTERNATIONAL DE COOPERATION SCIENTIFIQUE (PICS):
Synthesis of the ribosomal subunits in eukaryotic cells. Together with the Laboratoire de Biologie Moléculaire des Eucaryotes (LBME), Université de Toulouse, France

-
2004 - 2007
Marie Curie International Reintegration Grants (IRG) of the European Union: Chromosomal domains
collaboration
Hinrich Boeger, Molecular Cell & Developmental Biology, University of California Santa Cruz, USA.
Antonio Conconi, Département de microbiologie, Université de Sherbrooke, Canada.
Olivier Gadal, Laboratoire de Biologie Moléculaire Eucaryote, Université Paul Sabatier, Toulouse III, France.
Patrick Schultz, Institut de Génétique et de Biologie Moléculaire et Cellulaire, Illkirch, France.





