Research
| Fakultät für Biologie und Vorklinische Medizin |
| Institute of Plant Sciences |
| Cell Biology and Plant Biochemistry |
| Prof. Dr. Klaus Grasser - Research |
In eukaryotic cells, the large genomic DNA is packaged with histones and other proteins into chromatin. The compaction of the DNA provided by chromatin generally represses DNA-dependent processes such as the transcription of genes. During transcriptional elongation so-called transcript elongation factors facilitate transcript synthesis catalysed by RNA polymerase II. Using the Arabidopsis model system, we examine how these factors are involved in gene expression, development and plant responses to changing environmental conditions. In addition we are interested in the interplay with co-transcriptional mRNA processing and the mechanism of the nucleo-cytosolic transport of mRNAs. Towards these goals we employ approaches from the fields of molecular and cellular biology as well as biochemistry and genetics.
Transcript elongation factors
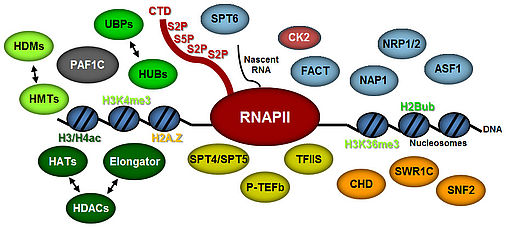
Transcript elongation by RNA polymerase II (RNAPII), once regarded as the simple extension of the initiated mRNA, is a complex and highly regulated phase of the transcription cycle. Many factors have been identified that contribute to the dynamic control of the elongation stage of transcription. There are transcript elongation factors (TEFs) that modulate the activity of RNAPII (green symbols in the scheme), while others facilitate the transcription through chromatin (blue and orange symbols), or covalently modify nucleosomal histones (i.e. methylation, acetylation, ubiquitination) within the transcribed regions (yellow symbols) (Van Lijsebettens and Grasser, 2014). Several TEFs including TFIIS, FACT, SPT4/SPT5 and PAF1C were found to associate with the elongating RNAPII (S2P, S5P phosphorylated) forming the transcript elongation complex (Antosz et al., 2017).
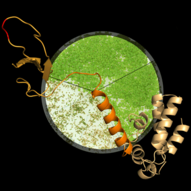
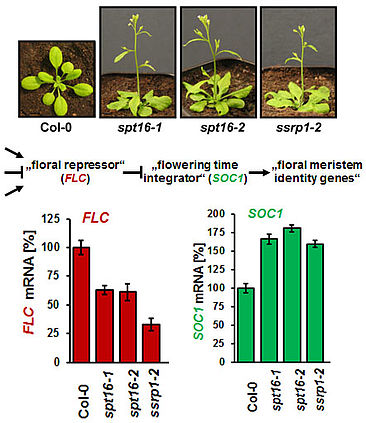
The Arabidopsis FACT (facilitates chromatin transcription) complex is composed of the SSRP1 and the SPT16 proteins and localises to the transcriptionally active euchromatin of the majority of cell types. The FACT histone chaperone is found to associate with the transcribed region of active genes in a transcription-dependent manner, suggesting that the FACT complex acts as transcript elongation factor assisting transcription in the chromatin context (Grasser, 2020). Arabidopsis mutant plants (ssrp1, spt16) that express reduced amounts of FACT subunits display various developmental defects including elevated number of smaller leaves, early bolting/flowering, and a bushy appearance due to reduced apical dominance. In addition, the architecture of mutant flowers is affected resulting in severely reduced seed production (Lolas et al., 2010). The early bolting of the mutant plants is associated with reduced transcript levels of the floral repressor FLC and consistently increased expression of SOC1.
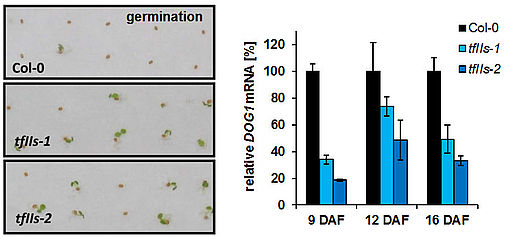
Seed germination analyses revealed that FACT contributes to the regulation of seed dormancy by promoting the expression of the DOG1 gene (Michl-Holzinger et al., 2019). Thus, FACT facilitates expression of DOG1 and FLC adjusting the crucial developmental transitions from seed dormancy to germination and from vegetative to reproductive development, respectively. Another study demonstrated that upon exposure to moderate high-light stress several genes encoding enzymes of the anthocyanin biosynthetic pathway are up-regulated to a lesser extent in ssrp1/spt16 compared to wild type plants. Accordingly, the mutant plants accumulate lower amounts of the purple anthocyanin pigments (Pfab et al., 2018). These findings indicate that FACT is a novel factor required for the accumulation of anthocyanins when the plants are exposed to environmental stress. The acidic region of the FACT subunit SPT16 is phosphorylated by protein kinase CK2 and the phosphorylation is required for establishing the correct nucleosome occupancy at transcriptional start sites of a subset of genes (Michl-Holzinger et al., 2022).

Another TEF that we are studying is TFIIS, an RNAPII-associated factor that can facilitate RNAPII transcription through sites that cause transient pausing or permanent arrest of transcribing RNAPII. Arabidopsis TFIIS can partially complement the phenotype of yeast cells lacking the endogenous TFIIS, indicating that Arabidopsis TFIIS plays a role in transcript elongation comparable to its yeast/mammalian counterparts. Mutant Arabidopsis plants develop normal, but the mutant seeds are severely affected in seed dormancy (Grasser et al., 2009). Thus, tfIIs seeds of fully elongated but still green siliques germinate efficiently, whereas comparable Col-0 seeds hardly germinate. The dormancy defect of tfIIs seeds is caused by reduced expression of the DOG1 gene encoding a potent regulator of seed dormancy (Mortensen and Grasser, 2014).
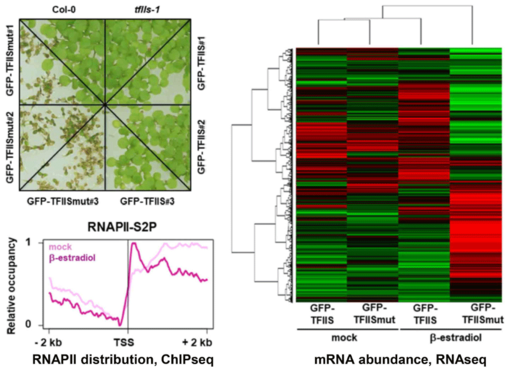
The elongation factor TFIIS stimulates the weak intrinsic transcript cleavage activity of RNAPII, which is required for efficient rescue of backtracked/arrested polymerase. A TFIIS mutant variant (TFIISmut) lacks the stimulatory activity to promote RNA cleavage, but instead efficiently inhibits unstimulated transcript cleavage by RNAPII. Constitutive expression of TFIISmut is lethal in Arabidopsis tfIIs plants. Induced, transient expression of TFIISmut in tfIIs plants provoked severe growth defects, transcriptomic changes and massive, transcription-related redistribution of elongating RNAPII within transcribed regions towards the transcriptional start site. The predominant site of RNAPII accumulation overlapped with the +1 nucleosome, suggesting that upon inhibition of RNA cleavage activity RNAPII arrest prevalently occurs at this position (Antosz et al., 2020). These findings suggest that polymerase backtracking/arrest frequently occurs in plant cells and RNAPII-reactivation is essential for correct transcriptional output and proper growth/development.
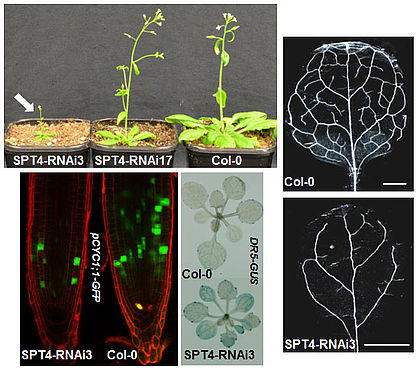
The SPT4 and SPT5 proteins interact in Arabidopsis cells forming the TEF termed SPT4/SPT5 that associates with RNAPII. SPT5 co-localises with elongating RNAPII in the euchromatin and is found in the transcribed regions of actively transcribed genes, but not of inactive genes. Down-regulation of SPT4 expression causes various vegetative and reproductive defects including severely impaired growth resulting from decreased cell proliferation. SPT4-RNAi plants show various auxin-signalling phenotypes (e.g. lateral root density, leaf venation) and a number of auxin-related genes are down-regulated. Consistent with the decreased expression of AUX/IAA genes the SPT4-RNAi plants show an enhanced auxin-response (Dürr et al., 2014). The perturbed development most likely is caused by transcript elongation defects in the SPT4-RNAi plants, as evident from RNAPII accumulation over the transcribed regions of genes.
These examples illustrate the important role of the regulation of transcript elongation in plant developmental processes that we intend to study in more detail in future research.
mRNA export factors
Various processing steps including 5´capping, splicing and 3´processing occur essentially during RNA polymerase II transcription, converting pre-mRNAs into mature mRNAs. Before mRNAs can be translated in the cytosol they have to be exported from the cell nucleus through nuclear pore complexes (NPCs). The regulated nucleo-cytosolic transport assures that only properly processed mRNAs are passed on to the cytosol for translation. Therefore, the export of mRNAs represents a critical step in gene expression. Accordingly, a variety of mRNA export factors is recruited to the mRNA co-transcriptionally that render the mature transcript export-competent (Ehrnsberger et al., 2019a). A central player in mRNA export is the THO/TREX complex, consisting of the THO core subunits that associate with additional factors including the UAP56 RNA helicase, ALY-/UIF-like proteins and MOS11 (Sørensen et al., 2017). Finally, adaptor proteins such as ALY recruit the soluble mRNA export receptor (ExR, which is unknown in plants) that mediates translocation of the export-competent mRNP through the NPC.
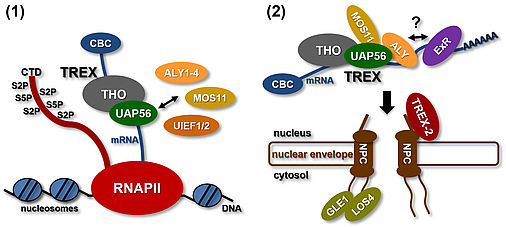
Arabidopsis UAP56 is a DEAD-box helicase and two neighbouring genes encode an identical protein that is ubiquitously expressed and localises primarily to the nucleoplasm in root and leaf cells. Recombinant UAP56 binds both to ssRNA and dsRNA, and it has RNA-stimulated ATPase activity as well as ATP-dependent RNA helicase activity resulting in the unwinding of dsRNA (Kammel et al., 2013). UAP56 interacts directly with ALY/UIEF proteins and MOS11, and therefore may be involved in the recruitment of these export factors.

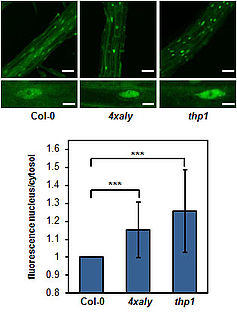
Export factors of the ALY family are more diversified in plants than in other organisms and Arabidopsis expresses four ALY proteins termed ALY1-4. ALY1 and ALY2 are closely related and likewise ALY3 and ALY4 share a high degree of amino acid sequence identity. In vitro RNA-binding assays demonstrate that in addition to the central RNA recognition motif (RRM) of ALY1 both the N- and C-terminal domains contribute to the RNA interactions. Plants defective in single ALY genes as well as double-mutant plants exhibit basically wild type appearance. However, quadruple mutant plants defective in the expression of all four ALY genes (4xaly) display various vegetative and reproductive abnormalities including small size, reduced root growth and decreased fertility (Pfaff et al., 2018). In line with this observation, mRNA export is reduced in 4xaly plants as evident from nuclear accumulation of polyadenylated mRNAs detected by in situ hybridisation with fluorescently labelled oligo(dT) probes. A second group of mRNA export factors termed UIEF1/2 cooperate with the ALY proteins to facilitate translocation of mRNAs through NPCs to the cytosol (Ehrnsberger et al., 2019b).
 |  |
These results demonstrate that proper plant development requires efficient nucleo-cytosolic transport of mRNAs, whose mechanism we intend to study in more detail in future experiments.
Architectural chromosomal proteins
HMGB proteins
High mobility group (HMG) proteins are a class of chromosomal non-histone proteins common to eukaryotic organisms. They are among the most abundant and ubiquitous proteins in the nucleus. HMG proteins act as architectural factors facilitating the proper control of various DNA dependent processes (transcription, recombination etc.). Higher plants express, depending on the species, one or two HMGA proteins and a variety of relatively abundant HMGB protein
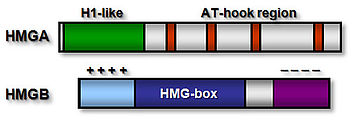
The HMGA proteins bind A/T-stretches of double-stranded DNA, while the HMGB proteins bind non-sequence-specifically to DNA. Plant HMGB proteins recognise various distorted DNA structures, and bend the DNA upon binding. The basic N-terminal domain enhances the general affinity of HMGB1 for linear DNA by direct contacts with DNA; conversely, the acidic C-terminal domain reduces the affinity to a comparable extent. The opposing effects of the N- and C-terminal domains appear to be balanced in HMGB1, resulting in similar affinity for linear DNA of full-length HMGB1 and the individual HMG-box domain. In maize HMGB1, the N- and C-terminal domains can interact, functionally 'neutralising' each other. In living cells, HMGA and HMGB proteins interact only transiently with chromatin, and accordingly, HMG proteins are highly dynamic within the cell nucleus, constantly scanning the nuclear space for binding sites.
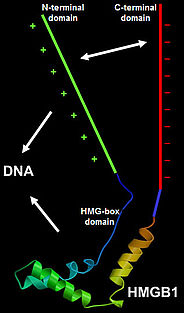
HMGB proteins are phosphorylated in plants and we could demonstrate that the maize HMGB1 and HMGB2/3 proteins, for instance, are differentially phosphorylated by protein kinase CK2 in vitro and in vivo. CK2 phosphorylates residues within the C-terminal acidic domains, which strengthens the intramolecular interaction between the basic N-terminal and the acidic C-terminal domains. The phosphorylation reduces the affinity of the proteins for linear DNA, but has only little influence on the recognition of DNA minicircles.
Many biological processes that involve regulated DNA transactions are dependent on protein-induced distortions in DNA structure brought about by DNA-bending proteins. These accessory factors bend the DNA locally, and stimulate the assembly of complex nucleoprotein structures required for biological function. The non-sequence-specific DNA-bending proteins can be recruited to their site of action by interactions with sequence-specific regulatory proteins. The HMGB proteins, for instance, can functionally interact with the maize transcription factor Dof2, stimulating its DNA-binding.
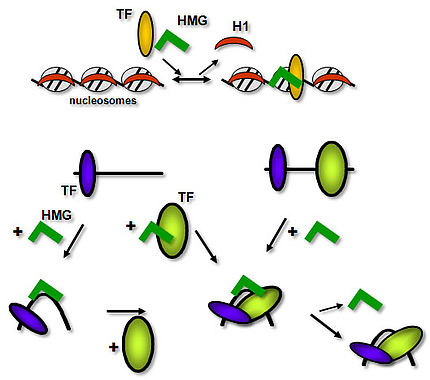
HMG proteins are considered “general” chromosomal proteins that may exert their function by exchanging with linker histone H1. In addition, HMG proteins functionally interact with transcription factors (TFs), facilitating their binding to target sites, and thereby they contribute to transcriptional regulation.
Other HMG-box proteins
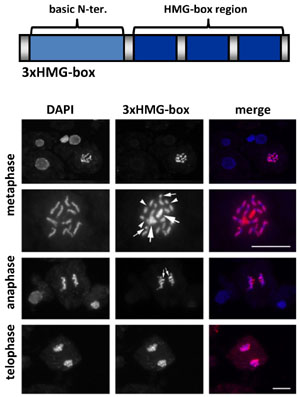
In addition to the HMGB proteins, the HMG-box domain is found in other plant DNA-binding proteins including SSRP1 (see transcript elongation factors) and ARID-HMG proteins that contain an AT-rich interaction domain (ARID) and an HMG-box domain. More recently, we have identified a fourth protein family containing three copies of the HMG-box domain in combination with an N-terminal basic domain and they were termed 3xHMG-box proteins. In contrast to other HMG-box proteins, the two Arabidopsis 3xHMG-box proteins are expressed in cells undergoing mitosis or meiosis, but not in interphase cells. They interact specifically with the condensed chromosomes during mitosis. 3xHMG-box2 generally associates with the condensed chromosomes, while 3xHMG-box1 preferentially binds rDNA loci.