Research
The primary scientific goal of the research group is to understand the relationship between protein motions and protein function. This is especially relevant for enzymes that have to undergo structural rearrangements to perform biological tasks. In the lab we focus on understanding the mechanism behind the bio-molecular complexes that play a role in the degradation of mRNA.
mRNA degradation
All organisms require a reliable mechanism to turn genes on and off. This regulation of gene expression underlies cellular processes ranging from the response to environmental signals to the development of multi-cellular organisms and cell-cell communication. Understandably, the cell tightly controls gene expression at every step from DNA to protein. Recent work has given new insights into these control mechanisms and revealed dedicated pathways that target mRNA for degradation, thereby efficiently turning genes off.
After export to the cytoplasm, mRNA is protected from degradation by a 5’ cap structure and a 3’ poly adenine tail. In the deadenylation dependent mRNA decay pathway, the poly(A) tail is gradually shortened by exonucleases. This ultimately attracts the degradation machinery that rapidly degrades the mRNA in both the 5’ to 3’ direction and in the 3’ to 5’ direction. Additional mechanisms, including the nonsense mediated decay pathway, bypass the need for deadenylation and can remove the mRNA from the transcriptional pool independently. Interestingly, the same enzymes are responsible for the actual degradation of the mRNA independent of the pathway taken (see figure).
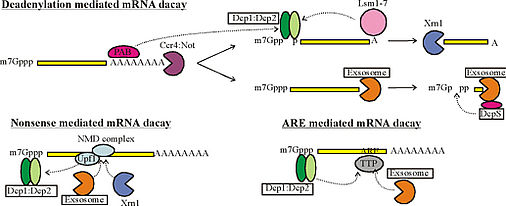
Figure 1: Simplified mechanism of mRNA decay. Dotted arrows indicate (indirect) interactions. Details of the pathways vary between different eukaryotes. PAB: poly-A binding protein.
Crystal structures for some of the complexes that play a role in mRNA decay have been solved, including the Dcp1:Dcp2 decapping complex, the DcpS scavenger decapping complex and the multi-component exosome complex. Our understanding of enzyme function is, however, limited to a static three dimensional fold of one of the many conformations that these proteins can adopt. To fully understand how molecular motions lead to catalytic activity, a complete picture of the protein dynamics is required. In addition, the catalytic activity of these enzymes must be tightly regulated to prevent premature degradation of mRNA and to ensure maximum activity as soon as an mRNA has been identified as a substrate. As such, intermolecular interactions that modulate catalytic activity, e.g. by restricting molecular motions, are of foremost interest.
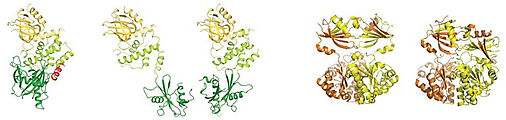
Figure 2: Examples of protein decapping enzymes that can adept different conformations. Left: the Dcp1:Dcp2 complex (She et. al. Mol Cell 2008, She et. al. Nat Struct Mol Biol. 2006; 2QKM, 2A6T). Right: DcpS in the open and closed state (Gu et. al. Mol Cell 2004; 1XMM, 1XML)
NMR Spectroscopy
Nuclear Magnetic Resonance (NMR) spectroscopy is the method of choice to detect and quantify biologically important motions in a near cellular environment. Using dedicated experiments, we can probe motions ranging from the pico- to nanoseconds time-scale up to the seconds time-scale, with atomic resolution. In addition, we can monitor how the molecular motions are affected by the interaction with adaptor complexes.
Traditionally, the application of NMR spectroscopy has been limited to complexes below 25 kDa in molecular weight. Recent advances have, however, extended this molecular weight regime into the hundreds of kDa and in favorable cases over 1 MDa. This makes the large complexes involved in mRNA decay amenable to detailed studies of molecular dynamics.
In summary, we hope to obtain a complete picture of protein dynamics and the modulation thereof. These aspects are fundamental to the understanding of the delicate balance between mRNA synthesis and degradation that allows the cell to regulate cellular processes by rapidly switching specific genes on or off.
Publications
CV
-
1993-1998: Chemistry Studies, Utrecht University, The Netherlands
-
1999-2003: PhD with Michael Sattler, EMBL Heidelberg, Germany
-
2003-2008: Postdoctoral fellow with Lewis Kay, University of Toronto, Canada
-
2008-2016: Group Leader (W2) at the MPI for Developmental Biology, Tübingen, Germany
-
2016-present: Professor of Biophysics (W3), Regensburg University, Germany
funding
The research in our group is supported by the ERC and the DFG via the Sonderforschungsbereich 960




