rDNA Chromatin and transcription
Chromatin consisting of DNA and associated proteins plays a central role in regulating protein access to DNA and influences essential processes such as transcription, replication, and DNA repair. Chromatin dynamics vary depending on the functional state of genomic regions.
We use Saccharomyces cerevisiae (yeast) as a model system because it is amenable to simple genetic manipulation and offers excellent biochemical accessibility. Our focus lies on two genomic loci: the RNA polymerase II–dependent PHO5 gene and the ribosomal DNA (rDNA) locus, which contains genes transcribed by RNA polymerases I and III. This enables us to perform comparative analyses of chromatin transcribed by all three eukaryotic RNA polymerases.
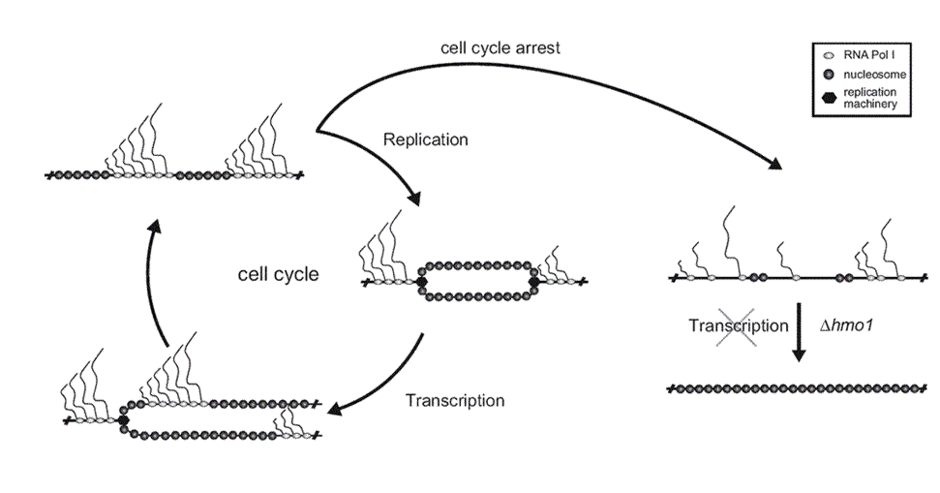
Of particular interest are the ribosomal RNA (rRNA) genes within the rDNA locus. In S. cerevisiae, 150 to 200 tandemly repeated transcription units are located on the right arm of chromosome XII. Each unit contains the 35S rRNA gene transcribed by RNA polymerase I and the 5S rRNA gene transcribed by RNA polymerase III. In actively dividing yeast cells, rRNA synthesis by RNA polymerase I accounts for approximately 70% of total transcription.
A fascinating aspect of our research is the existence of two populations of 35S rRNA genes: actively transcribed genes and transcriptionally inactive genes that adopt distinct chromatin structures. While actively transcribed rRNA genes are largely devoid of nucleosomes and bound by the HMG-box protein Hmo1, maintaining a fraction of inactive 35S rRNA genes is crucial for genomic stability under certain conditions.
Our goal is to develop a comprehensive molecular description of chromatin in different transcriptional states. To this end, we analyze chromatin changes during transcription and investigate the mechanisms involved in the establishment, maintenance, and remodeling of chromatin.
