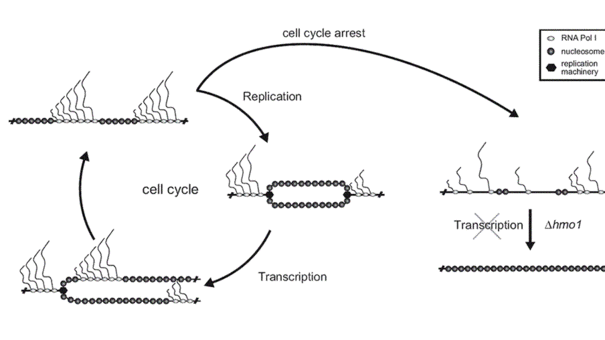
Recoding mechanisms during RNA translation
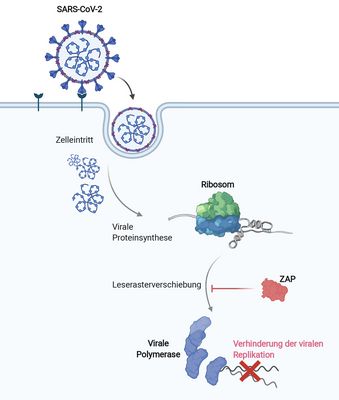
In viruses or cellular genes, certain RNAs can be read in alternative ways during translation, a process known as recoding. However, it remains unclear how exactly recoding is regulated by host or viral factors. A precise understanding of recoding and its regulation may therefore be key to developing new RNA-based. Our team has identified several viral RNA interaction partners that disrupt the synthesis of the SARS-CoV-2 polyprotein or HIV-1 replication. Building on this, my group investigates how these proteins dynamically interact with structured RNAs and the host’s translational machinery during translation. Identifying peptide-based or synthetic molecules that can specifically interact with viral RNA elements will help to understand how modulation of RNA can be used as an effective antiviral strategy.
Translational regulation during infection
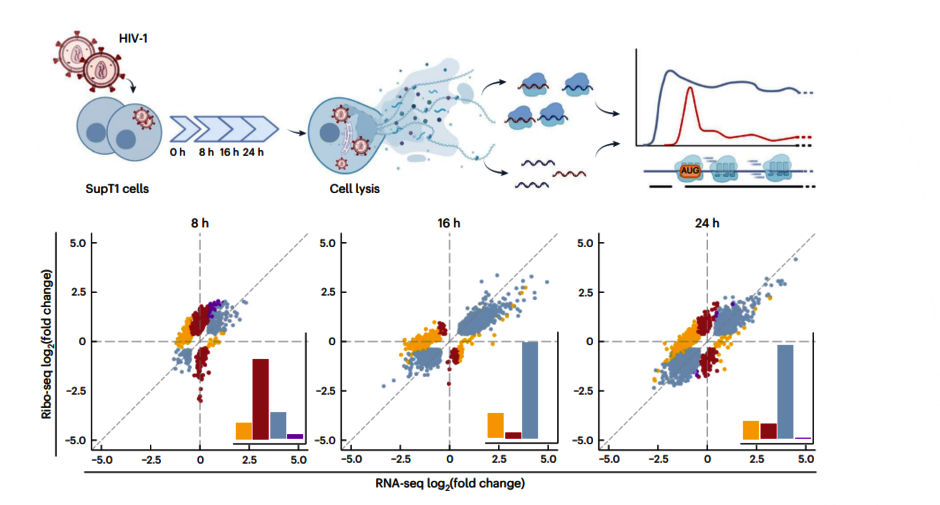
Changes in the environment, cellular differentiation, and infections can influence mechanisms of gene expression and thereby alter the cellular proteome in unexpected ways. Nevertheless, it is often unclear to what extent human cells actually employ non-standard translation events, for example during an infection.
To better understand translational processes and the role of RNA molecules in these processes, we use state-of-the-art RNA analytics such as ribosome profiling and deep sequencing. Our focus is on the dynamics of non-standard translational mechanisms and the potential role of cellular factors in this interplay. Ultimately, we aim to achieve a more comprehensive understanding of the interaction between host and pathogen gene expression.
Single molecule analysis of RNA complexes
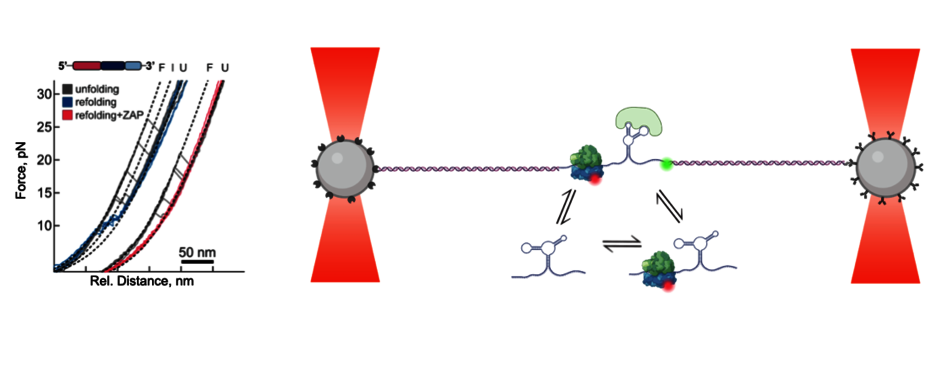
RNAs can form complex three-dimensional structures and interact with other regulatory elements such as ncRNAs, small molecules, and proteins to alter the function of the information encoded in the primary sequence of mRNA. How RNA structures and regulatory elements control alternative translation events is not yet fully understood. A key question we address is: “To what extent do the strength of RNA base pairing and the conformational dynamics of the structure define the propensity of ribosomes to shift into an alternative reading frame?” Using state-of-the-art single molecule and ensemble analysis tools, we investigate how trans-acting factors alter RNA structure. The tools we have developed serve as a starting point for the development of potent and specific modulators of frameshifting.